
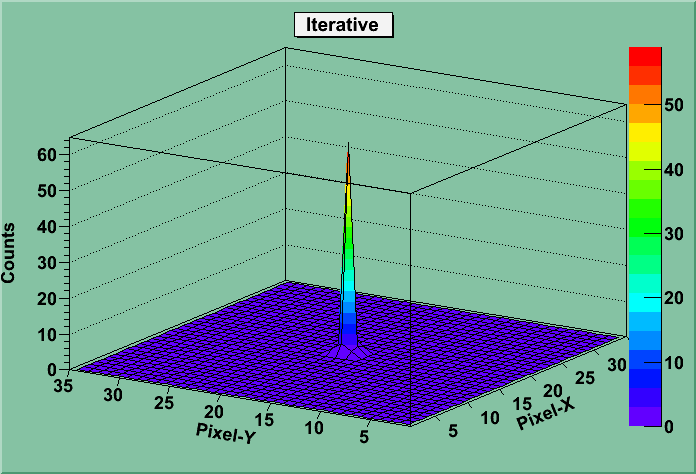
The following several columns give the peak position (in ppm), the scalar coupling (in Hz), and the R 2 (in Hz), with the integer suffix indicating the dimension for each parameter. If applicable, the F-test p-value will be given next.
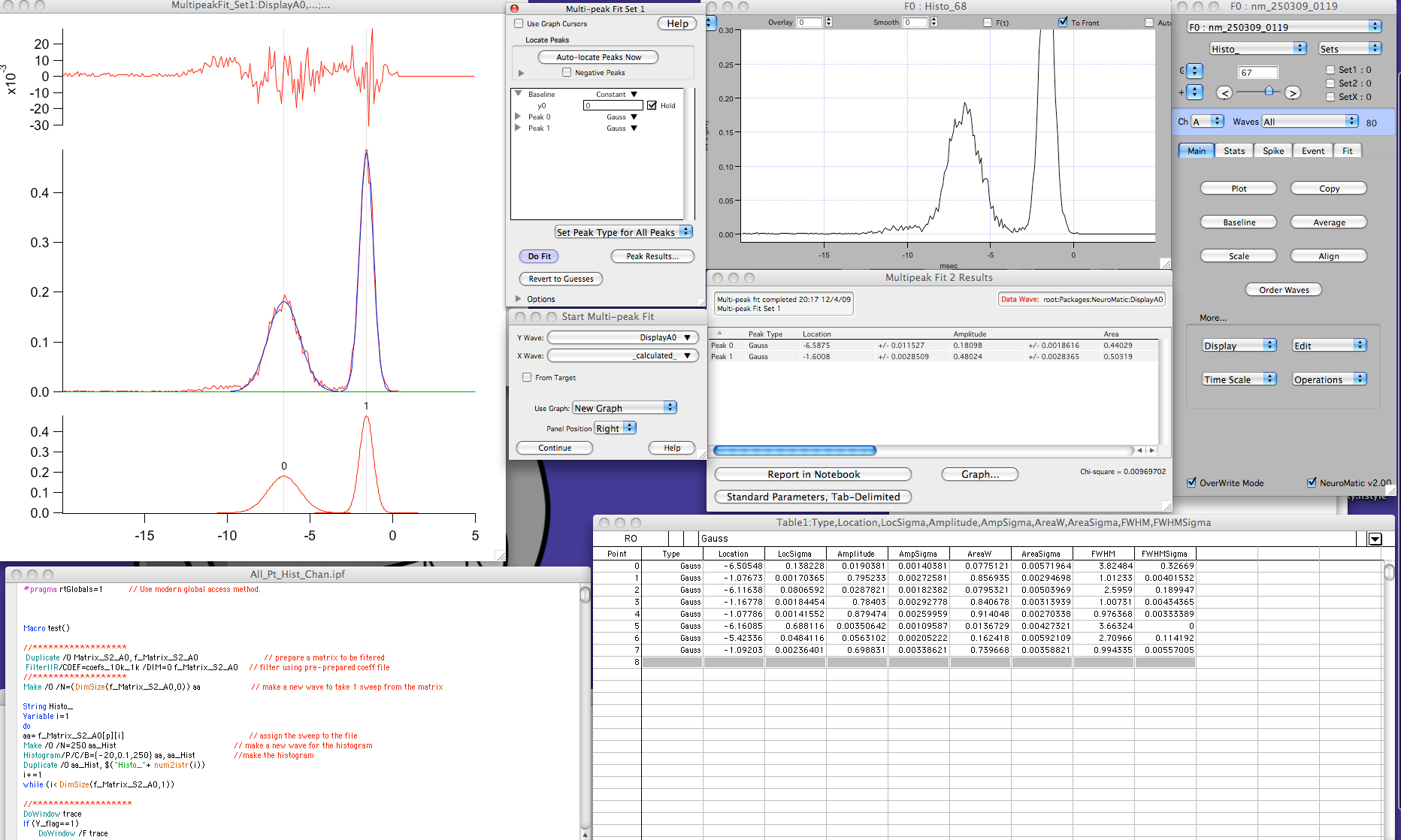
The first couple columns of that table give the peak number and fit iteration at which the peak was added. To convert that into a more user-friendly table, use the param_list_to_peak_df function. The fit_peak_iter function returns a list of fits, one for each iteration. For the first iteration, the search is terminated after the third peak fails to fall below the p-value cutoff, which is specified by f_alpha and defaults to 0.001. Adding the second peak involves addition of just 3 parameters (2 omega0 and 1 m0), because several of the parameters are shared with the first peak (2 r2 and 1 scalar coupling). For the first peak added, this is very low, indicating that having a peak at that position is much better than assuming otherwise. The probability of the observed improvement to the fit happening at random (according to an F-test) is given by the p-value. Adding the first peak involves addition of 6 parameters to the model (2 omega0, 2 r2, 1 m0, and 1 scalar coupling). (By contrast double square brackets,], return individual items from the list that are not enclosed in a list data structure.) Theiter_max argument infit_peak_iter` specifies the number of iterations to perform.įitNMR outputs text describing what the peak fitting algorithm is doing. To get a list of just one spectrum, use single square brackets,, which return another list. In this case, we will only do the fitting on the first spectrum. It takes a list of spectra, which we previously read into spec_list``. We will start by running three iterations of the iterative peak fitting method. header: The raw data from the 512-value NMRPipe header.fheader: A matrix of header values associated each dimension.ppm: A list of chemical shifts for each dimension stored in numeric format.The chemical shifts are stored in character format in the dimension names ( dimnames) of the array. int: A multidimensional array with the intensities of the spectrum, which in this case is just a 2D matrix.
Each spectrum is itself a list with four named components: This shows that there is a list of two spectra. attr(*, "names")= chr "FDMAGIC" "FDFLTFORMAT" "FDFLTORDER" "". $ : chr "QUADFLAG" "CAR" "CENTER" "FTSIZE". * Tests the aggregated plate statistics method using the values between the indices of Source Link DocumentReturns the maximum of the available values Usageįrom source file. /** IntroductionIn this page you can find the example usage for 3.scriptive DescriptiveStatistics getMax.


 0 kommentar(er)
0 kommentar(er)
